- Algorithm Step 1: Reliability of an alignment edge Step 2: Maximum expected accuracy Step 3: Probabilistic Consistency Transformation Step 4: Computation of guide tree Step 5: Compute MSA
- See also
- References
- External links
Algorithm
The following describes the basic outline of the ProbCons algorithm.[3]
Step 1: Reliability of an alignment edge
For every pair of sequences compute the probability that letters  and
and  are paired in
are paired in  an alignment that is generated by the model.
an alignment that is generated by the model.

(Where 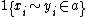 is equal to 1 if
is equal to 1 if  and
and  are in the alignment and 0 otherwise.)
are in the alignment and 0 otherwise.)
Step 2: Maximum expected accuracy
The accuracy of an alignment  with respect to another alignment
with respect to another alignment  is defined as the number of common aligned pairs divided by the length of the shorter sequence.
is defined as the number of common aligned pairs divided by the length of the shorter sequence.
Calculate expected accuracy of each sequence:
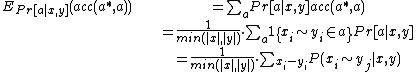
This yields a maximum expected accuracy (MEA) alignment:
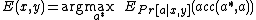
Step 3: Probabilistic Consistency Transformation
All pairs of sequences x,y from the set of all sequences  are now re-estimated using all intermediate sequences z:
are now re-estimated using all intermediate sequences z:
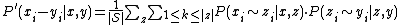
This step can be iterated.
Step 4: Computation of guide tree
Construct a guide tree by hierarchical clustering using MEA score as sequence similarity score. Cluster similarity is defined using weighted average over pairwise sequence similarity.
Step 5: Compute MSA
Finally compute the MSA using progressive alignment or iterative alignment.
See also
- Sequence alignment software
- Clustal
- MUSCLE
- AMAP
- T-Coffee
- Probalign
References
2. ^{{Cite book|title=Multiple Sequence Alignment Methods|volume = 1079|last=Roshan|first=Usman|date=2014-01-01|publisher=Humana Press|isbn=9781627036450|editor-last=Russell|editor-first=David J|series=Methods in Molecular Biology|pages=147–153|language=English|doi=10.1007/978-1-62703-646-7_9|pmid = 24170400|chapter = Multiple Sequence Alignment Using Probcons and Probalign}}
3. ^Lecture "Bioinformatics II" at University of Freiburg
External links
- {{Official website|http://probcons.stanford.edu/}}
- Ricci decomposition
- Ricci-flat
- Ricci flat
- Ricci-flat manifold
- Ricci-flow
- Ricci flow
- Ricci, Matteo
- Riccioli (crater)
- Riccioli crater
- Ricciotto Canudo
- Ricci scalar
- Ricci scalar curvature
- Ricci tensor
- Riccius (crater)
- Ricciutello
- Ricco (crater)
- Ricco Groß
- Ricco's law
- Ric Dagenais
- Rice
- Rice and curry
- Rice, Anne
- Rice ball
- Rice Belt
- Ricebird