侧锋
笔管倾斜,行笔时,笔尖在笔道的一边。侧锋富于变化。
侧锋Cefeng
用笔法之一。笔管倾斜,行笔时,笔尖在笔道的一边,侧锋富于变化。石涛说:“腕受正,则中直藏锋,腕受仄,则欹斜尽致。”
侧锋
中国绘画中笔锋的锋法之一。又称偏锋或卧锋。笔管倾斜,笔端、笔身、有时乃至笔根同时着纸。行笔时,笔尖在笔道一侧,笔锋露而不藏。侧锋用笔可绘出较粗宽的墨线,笔道中间富有虚实多变的墨色或飞白,可显示出钝重、粗旷而丰润的艺术效果。山水画中的皴法,多用侧锋运笔。可因行笔的疾徐、腕力的轻重、水墨之多少的不同,使笔画出现墨色各异的变化,还可出现笔道一边齐、一边毛的特有效果。
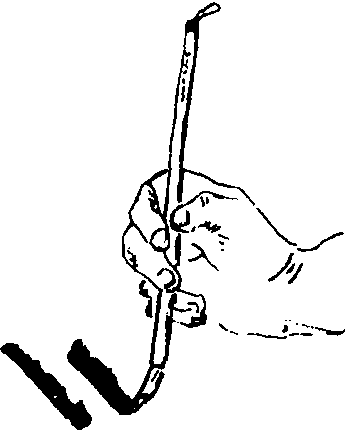
侧锋