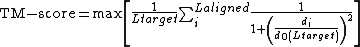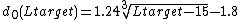- The equation
- See also
- References
- External links
In bioinformatics, the template modeling score or TM-score is a measure of similarity between two protein structures with different tertiary structures. The TM-score is intended as a more accurate measure of the quality of full-length protein structures than the often used RMSD measure. The TM-score indicates the difference between two structures by a score between 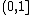 , where 1 indicates a perfect match between two structures (thus the higher the better).[1] Generally scores below 0.20 corresponds to randomly chosen unrelated proteins whereas structures with a score higher than 0.5 assume roughly the same fold.[2]
, where 1 indicates a perfect match between two structures (thus the higher the better).[1] Generally scores below 0.20 corresponds to randomly chosen unrelated proteins whereas structures with a score higher than 0.5 assume roughly the same fold.[2]
A quantitative study
[3]shows that proteins of TM-score = 0.5 have a posterior probability of 37% in the same CATH topology family and of 13% in the same SCOP fold family. The probabilities increase rapidly when TM-score > 0.5. The TM-score is designed to be independent of protein lengths. The Global Distance Test (GDT) algorithm, and its GDT TS score to represent "total score", is another measure of similarity between two protein structures with known amino acid correspondences (e.g. identical amino acid sequences) but different tertiary structures.[4]
The equation
where  and
and 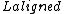 are the lengths of the target protein and the aligned region respectively.
are the lengths of the target protein and the aligned region respectively.  is the distance between the
is the distance between the 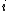 th pair of residues and
th pair of residues and
is a distance scale that normalizes distances.
See also
- RMSD — a different structure comparison measure
- GDT — a different structure comparison measure
- LCS — a different structure comparison measure
References
2. ^{{cite journal |author=Zhang Y and Skolnick J |title=TM-align: a protein structure alignment algorithm based on the TM-score |journal=Nucleic Acids Res |volume=33 | issue=7 |pages=2302–2309 |year=2005 |pmid=15849316 |doi=10.1093/nar/gki524 |pmc=1084323}}
3. ^{{cite journal |author=Xu J and Zhang Y |title=How significant is a protein structure similarity with TM-score = 0.5? |journal=Bioinformatics |volume=26 | issue=7 |pages=889–895 |year=2010 |pmid=20164152 |pmc=2913670 |doi=10.1093/bioinformatics/btq066}}
4. ^{{cite journal |author=Zemla A |year=2003 |title=LGA: A method for finding 3D similarities in protein structures |journal=Nucleic Acids Research |volume=31 |issue=13 |pages=3370–3374 |pmid=12824330 |pmc=168977 |doi=10.1093/nar/gkg571}}
External links
- TM-score webserver — by the Yang Zhang research group. Calculates TM-score and supplies source code.
- GDT and LGA description services and documentation on structure comparison and similarity measures.
- Colegio Nacional de Buenos Aires
- Colegio Nacional de Ushuaia
- Colegio Nacional Enrique Nvó Okenve
- Colegio Nueva Granada
- Colegio Pestalozzi (Argentina)
- Colegio San Ignacio de Loyola (San Juan)
- Colegio Tarbut
- Coleg Trefeca
- Coleham Pumping Station
- Coleharbor
- Coleharbor, ND
- Coleharbor, North Dakota
- Cole Harbour, Nova Scotia
- Cole Hauser
- Cole High School
- Colehill
- Colel Cab
- Coleman
- Coleman Alexander Young
- Coleman A. Young
- Coleman A. Young International Airport
- Colemanballs
- Coleman Coliseum
- Coleman Company
- Coleman Company, Inc.